Learning protein constitutive motifs from sequence data
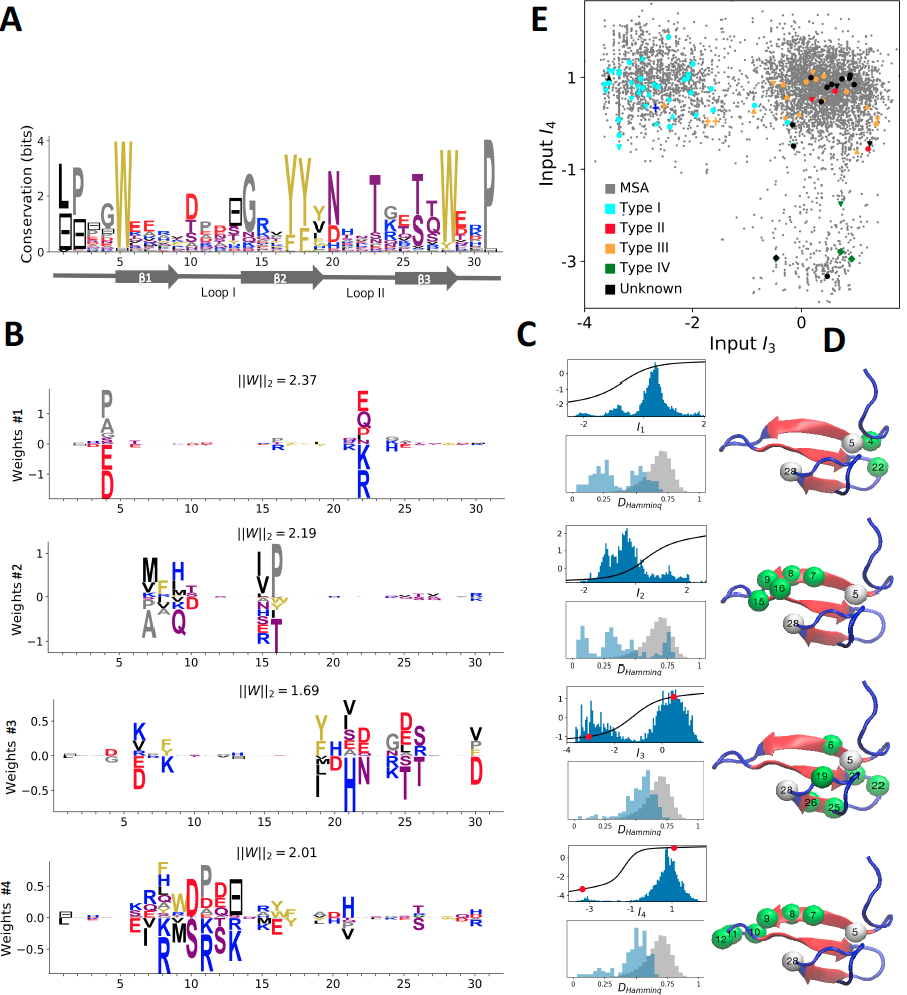
Statistical analysis of evolutionary-related protein sequences provides insights about their structure, function, and history. We show that Restricted Boltzmann Machines (RBM), designed to learn complex high-dimensional data and their statistical features, can efficiently model protein families from sequence information. We apply RBM to two protein domains, Kunitz and WW, and to synthetic lattice proteins for benchmarking. The features inferred by the RBM can be biologically interpreted in terms of structural modes, including residue-residue tertiary contacts and extended secondary motifs ($\alpha$-helix and $\beta$-sheet), of functional modes controlling activity and ligand specificity, or of phylogenetic identity. In addition, we use RBM to design new protein sequences with putative properties by composing and turning up or down the different modes at will. Our work therefore shows that RBM are a versatile and practical tool to unveil and exploit the genotype-phenotype relationship for protein families.
PDF Abstract