Skin Lesion Segmentation: U-Nets versus Clustering
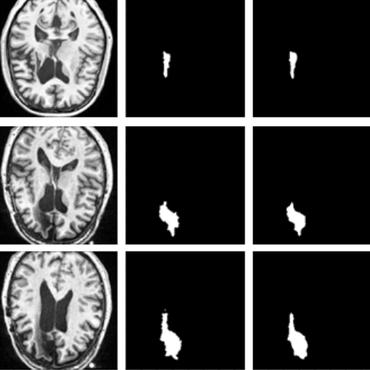
Many automatic skin lesion diagnosis systems use segmentation as a preprocessing step to diagnose skin conditions because skin lesion shape, border irregularity, and size can influence the likelihood of malignancy. This paper presents, examines and compares two different approaches to skin lesion segmentation. The first approach uses U-Nets and introduces a histogram equalization based preprocessing step. The second approach is a C-Means clustering based approach that is much simpler to implement and faster to execute. The Jaccard Index between the algorithm output and hand segmented images by dermatologists is used to evaluate the proposed algorithms. While many recently proposed deep neural networks to segment skin lesions require a significant amount of computational power for training (i.e., computer with GPUs), the main objective of this paper is to present methods that can be used with only a CPU. This severely limits, for example, the number of training instances that can be presented to the U-Net. Comparing the two proposed algorithms, U-Nets achieved a significantly higher Jaccard Index compared to the clustering approach. Moreover, using the histogram equalization for preprocessing step significantly improved the U-Net segmentation results.
PDF Abstract